Fitting
Note
If some code blocks are not readable, expand the documentation window
Preparing to fit data
To fit some data you must first load some data, activate one or more data sets, send those data sets to fitting, and select a model to fit to each data set.
Instructions on how to load and activate data are in the section Loading Data.
SasView can fit data in one of three ways:
- in Single fit mode - individual data sets are fitted independently one-by-one
- in Simultaneous fit mode - multiple data sets are fitted simultaneously to the same model with/without constrained parameters (this might be useful, for example, if you have measured the same sample at different contrasts)
- in Batch fit mode - multiple data sets are fitted sequentially to the same model (this might be useful, for example, if you have performed a kinetic or time-resolved experiment and have lots of data sets!)
Selecting a model
The models in SasView are grouped into categories. By default these consist of:
- Cylinder - cylindrical shapes (disc, right cylinder, cylinder with endcaps etc)
- Ellipsoid - ellipsoidal shapes (oblate,prolate, core shell, etc)
- Parallelepiped - as the name implies
- Sphere - spheroidal shapes (sphere, core multishell, vesicle, etc)
- Lamellae - lamellar shapes (lamellar, core shell lamellar, stacked lamellar, etc)
- Shape-Independent - models describing structure in terms of density correlation functions, fractals, peaks, power laws, etc
- Paracrystal - semi ordered structures (bcc, fcc, etc)
- Structure Factor - S(Q) models
- Plugin Models - User-created (custom/non-library) Python models
Use the Category drop-down menu to chose a category of model, then select a model from the drop-down menu beneath. A graph of the chosen model, calculated using default parameter values, will appear. The graph will update dynamically as the parameter values are changed.
You can decide your own model categorizations using the Category Manager.
Once you have selected a model you can read its help documentation by clicking on the Description button to the right.
Interaction Models
Structure factor S(Q) models can be combined with many form factor P(Q) models in the other categories to generate what SasView calls “interaction models” (previously “product models”). The combination can be done by one of two methods, but how they behave is slightly different.
The first, most straightforward, method is simply to use the S(Q) drop-down in the FitPage:
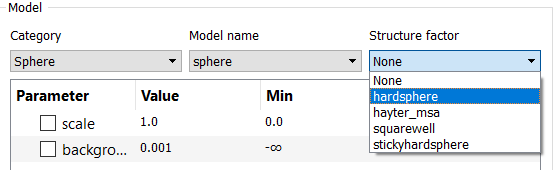
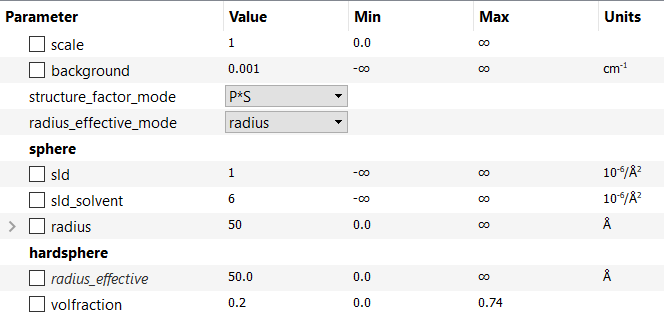
This example would then generate an interaction model with the following parameters:
The other method is to use the Sum|Multi(p1,p2) tool under Fitting > Plugin Model Operations:
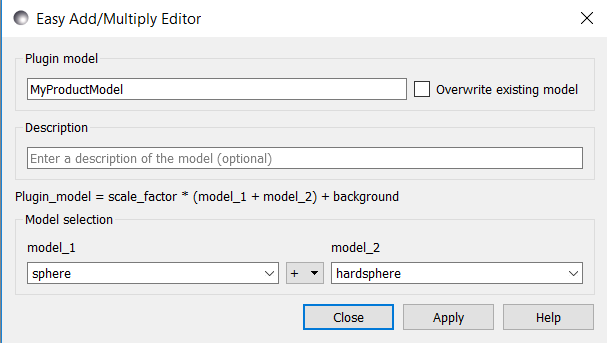
This creates an interaction model with the following parameters:
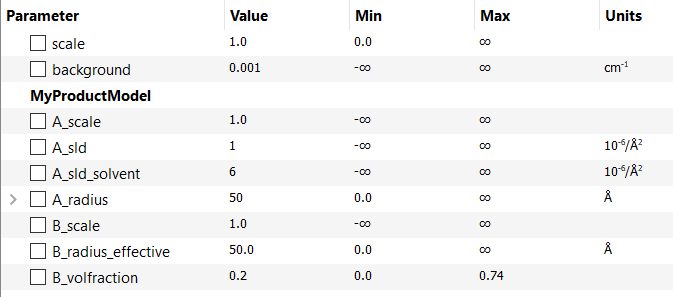
As can be seen, the second method has produced an interaction model with an extra parameter: radius_effective. This is the radial distance determining the range of the \(S(Q)\) interaction and may, or may not, be the same as the radius, in this example, depending on the concentration of the system. In other systems, radius_effective may depend on the particle form (shape). SasView offers the flexibility to automatically constrain (tie) some of these parameters together so that, for example, radius_effective = radius. See Sum|Multi(p1,p2).
Also see Fitting Models with Structure Factors for further information.
Mixture Models
SasView “mixture models” (previously called “sum models”) are summations of form factor models, or even of form factor models and an “interaction model” (see above), and are used to describe mixed-phase systems where the scattering is proportional to the volume fraction of each contributing phase.
Show 1D/2D
Models are normally fitted to 1D (ie, I(Q) vs Q) data sets, but some models in SasView can also be fitted to 2D (ie, I(Qx,Qy) vs Qx vs Qy) data sets.
NB: Magnetic scattering can only be fitted in SasView in 2D.
To activate 2D fitting mode, click the Show 2D button on the Fit Page. To return to 1D fitting model, click the same button (which will now say Show 1D).
Category Manager
To change the model categorizations, either choose Category Manager from the View option on the menubar, or click on the Modify button on the Fit Page.
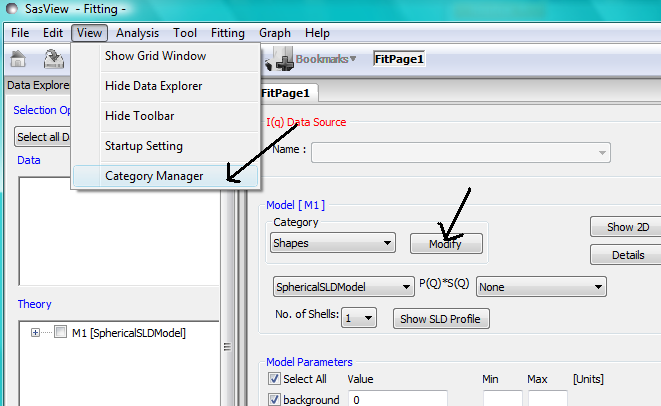
The categorization of all models except the user supplied Plugin Models can be reassigned, added to, and removed using Category Manager. Models can also be hidden from view in the drop-down menus.
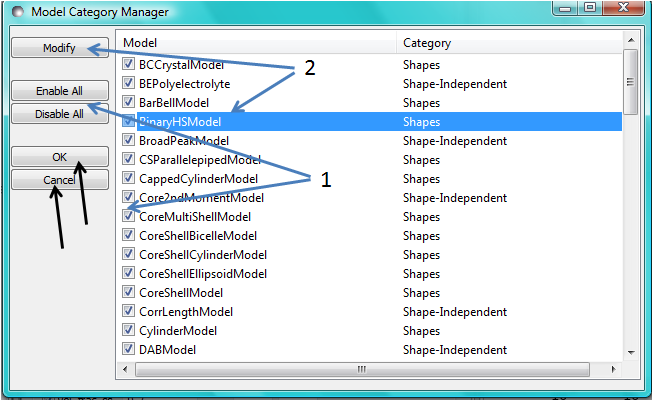
Changing category
To change category, highlight a model in the list by left-clicking on its entry and then click the Modify button. Use the Change Category panel that appears to make the required changes.
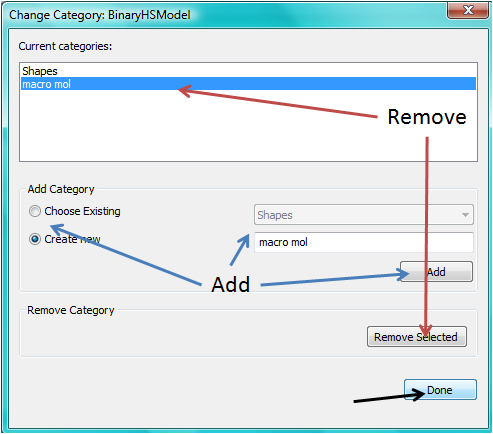
To create a category for the selected model, click the Add button. In order to delete a category, select the category name and click the Remove Selected button. Then click Done.
Showing/hiding models
Use the Enable All / Disable All buttons and the check boxes beside each model to select the models to show/hide. To apply the selection, click Ok. Otherwise click Cancel.
NB: It may be necessary to change to a different category and then back again before any changes take effect.
Model Functions
For a complete list of all the library models available in SasView, see the Model Documentation .
It is also possible to add your own models.
Adding your own Models
There are essentially four ways to generate new fitting models for SasView:
- Using the SasView Sum|Multi(p1,p2) dialog to sum/multiply together two existing models in the model library (best for beginners - provided the required models are in the model library!)
- Using the SasView New Plugin Model helper dialog (aimed at those with little programming experience but works best for relatively simple models)
- By copying/editing an existing model (this can include models generated by the New Plugin Model dialog) in the Python Shell-Editor Tool or Advanced Plugin Editor (suitable for all use cases)
- By writing a model from scratch outside of SasView (only recommended for experienced Python users)
In the last two cases, please read the guidance on Writing a Plugin Model before proceeding.
To be found by SasView your model must reside in the *~\.sasview\plugin_models* folder.
Plugin Model Operations
From the Fitting option in the menu bar, select Plugin Model Operations
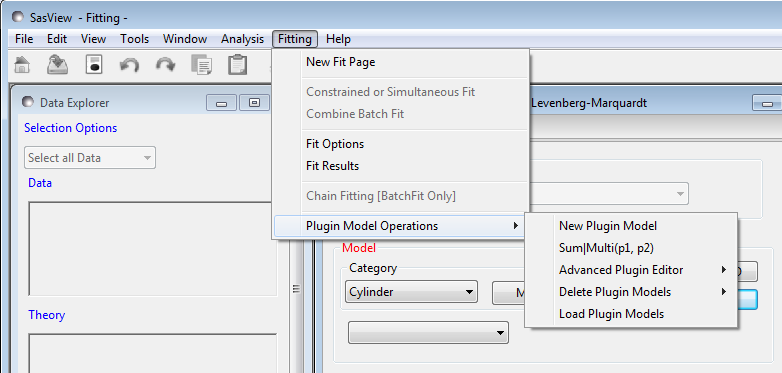
and then one of the sub-options
- New Plugin Model - to create a plugin model template with a helper dialog
- Sum|Multi(p1,p2) - to create a plugin model by summing/multiplying two existing models in the model library
- Advanced Plugin Editor - to create/edit a plugin model in a Python shell
- Delete Plugin Models - to delete a plugin model
- Load Plugin Models - to (re-)load plugin models
New Plugin Model
Relatively straightforward models can be programmed directly from the SasView GUI using the New Plugin Model Function.
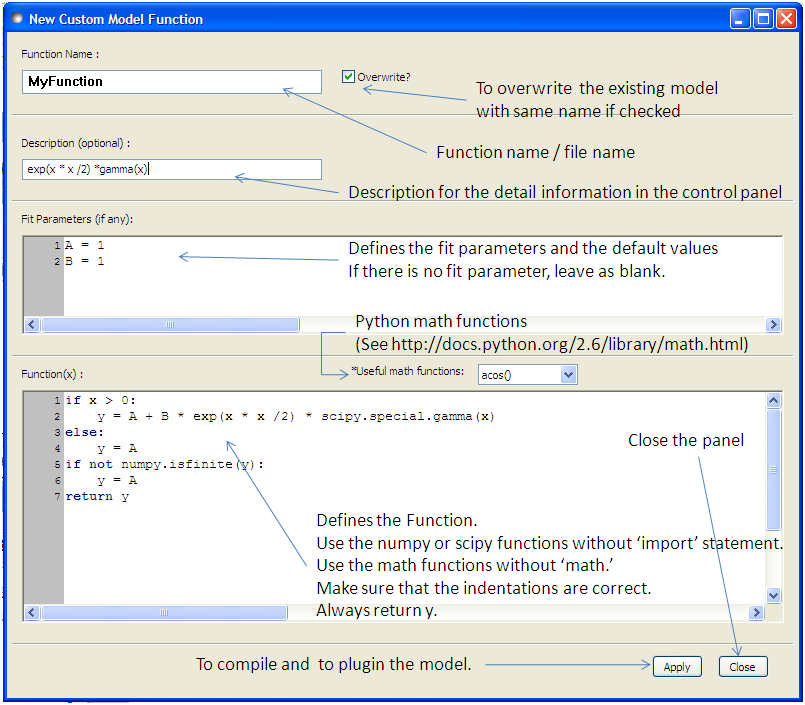
When using this feature, be aware that even if your code has errors, including syntax errors, a model file is still generated. When you then correct the errors and click ‘Apply’ again to re-compile you will get an error informing you that the model already exists if the ‘Overwrite’ box is not checked. In this case you will need to supply a new model function name. By default the ‘Overwrite’ box is checked.
A model file generated by this option can be viewed and further modified using the Advanced Plugin Editor .
Note that the New Plugin Model Feature currently does not allow for parameters to be polydisperse. However they can be edited in the Advanced Editor.
SasView version 4.2 made it possible to specify whether a plugin created with the New Plugin Model dialog is actually a form factor P(Q) or a structure factor S(Q). To do this, simply add one or other of the following lines under the import statements.
For a form factor:
form_factor = True
or for a structure factor:
structure_factor = True
If the plugin is a structure factor it is also necessary to add two variables to the parameter list:
parameters = [
['radius_effective', '', 1, [0.0, numpy.inf], 'volume', ''],
['volfraction', '', 1, [0.0, 1.0], '', ''],
[...],
and to the declarations of the functions Iq and Iqxy::
def Iq(x , radius_effective, volfraction, ...):
def Iqxy(x, y, radius_effective, volfraction, ...):
Such a plugin should then be available in the S(Q) drop-down box on a FitPage (once a P(Q) model has been selected).
Sum|Multi(p1,p2)
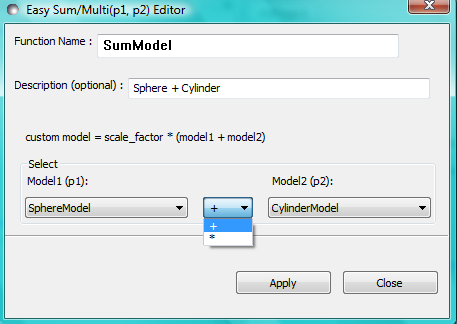
This option creates a custom Plugin Model of the form:
Plugin Model = scale_factor * {(scale_1 * model_1) +/- (scale_2 * model_2)} + background
or:
Plugin Model = scale_factor * (model1 * model2) + background
In the Easy Sum/Multi Editor give the new model a function name (which will appear in the list of plugin models on the FitPage) and brief description (to appear under the Details button on the FitPage). The model name must not contain spaces (use underscores to separate words if necessary) and if it is longer than ~40 characters the name will not display in full in the list of models. Now select two built-in models, as Model1 (p1) and Model2 (p2), and the required operator, ‘+’ or ‘*’ between them. Finally, click the Apply button to generate and test the model, and then click Close.
Any changes to a plugin model generated in this way only become effective after it is re-selected from the plugin models drop-down menu on the FitPage. If the model is not listed you can force a recompilation of the plugins by selecting Fitting > Plugin Model Operations > Load Plugin Models.
Warning
SasView versions 4.2.x, 5.0.0 and 5.0.1 The Easy Sum/Multi Editor dialog should not be used to combine a plugin model with a built-in model, or to combine two plugin models. The operation will appear to work in 4.2.x but may generate a faulty plugin model. In 5.0.0 the operation will fail (generating an error message in the Log Explorer). Whilst in 5.0.1 the operation has been blocked.
If you need to generate a plugin model from more than two built-in models, please read the sub-sections Model Structure and Combining more than two models below.
Model Structure
SasView version 4.2 introduced a much simplified and more extensible structure for plugin models generated through the Easy Sum/Multi Editor. For example, the code for a combination of a sphere model with a power law model now looks like this:
from sasmodels.core import load_model_info
from sasmodels.sasview_model import make_model_from_info
model_info = load_model_info('sphere+power_law')
model_info.name = 'MyPluginModel'
model_info.description = 'sphere + power_law'
Model = make_model_from_info(model_info)
To change the models or operators contributing to this plugin it is only necessary to edit the string in the brackets after load_model_info, though it would also be a good idea to update the model name and description too!!!
The model specification string can handle multiple models and combinations of operators (‘+’ or ‘*’) which are processed according to normal conventions. Thus ‘model1+model2*model3’ would be valid and would multiply model2 by model3 before adding model1. In this example, parameters in the FitPage would be prefixed A (for model2), B (for model3) and C (for model1). Whilst this might appear a little confusing, unless you were creating a plugin model from multiple instances of the same model the parameter assignments ought to be obvious when you load the plugin.
This streamlined approach to building complex plugin models from existing library models, or models available on the Model Marketplace, also permits the creation of P(Q)*S(Q) plugin models, something that was not possible in earlier versions of SasView. Also see Interaction Models above.
Note
Interaction Models
When the Easy Sum/Multi Editor creates a P(Q)*S(Q) model it will use the * symbol like this:
sphere*hardsphere
However, it is probably advisable to edit the model file and use the @ symbol instead, for example:
sphere@hardsphere
This is because * and @ confer different behaviour on the model
- with @ - the radius and volume fraction in the S(Q) model are constrained to have the same values as the radius and volume fraction in the P(Q) model.
- with * - the radii and volume fractions in the P(Q) and S(Q) models are unconstrained.
Warning
If combining P(Q) models with S(Q) models, particularly if combining multiple instances of such models (eg, \((P(Q)_1\) * \(S(Q)_1\)) + \((P(Q)_2\) * \(S(Q)_2)\) or similar), pay careful attention to the behaviour of the scale and volume fraction parameters and test your model thoroughly, preferably on well-characterised data.
Combining more than two models
If you need generate a plugin model from more than two other models, it is tempting to think that the way to do so is simply to use the Easy Sum/Multi Editor dialog to combine the first two models into a plugin, then generate a new plugin using that first plugin as one of the selected models, combine it with the third model, and repeat as required.
This does not currently work properly (although it may appear to in SasView 4.2.x).
Instead, use the Easy Sum/Multi Editor dialog to combine the first two models, then navigate to the plugin folder (~\.sasview\plugin_models on Windows) and open the plugin Python file (eg, MyPluginModel.py) in a text editor.
Now edit the Python to specify all the models to contribute to the expanded plugin (the text string in the brackets after load_model_info). Make sure you specify the model names correctly, including any capitalisation (if in doubt use the model name dropdown on a FitPage)! Finally, update the model name and description, and save the file.
So, as an example, one could take the MyPluginModel example in the preceding section, change it to:
from sasmodels.core import load_model_info
from sasmodels.sasview_model import make_model_from_info
model_info = load_model_info('power_law + fractal + gaussian_peak + gaussian_peak')
model_info.name = 'MyBigPluginModel'
model_info.description = 'For fitting pores in crystalline framework'
Model = make_model_from_info(model_info)
and re-save it as MyBigPluginModel.py. When loaded into a FitPage, the parameters for the four models in the load_model_info string are then all present and prefixed by A_, B_, C_, and D_, respectively.
Advanced Plugin Editor
Selecting this option shows all the plugin models in the plugin model folder, on Windows this is
C:\Users\{username}\.sasview\plugin_models
You can edit, modify, and save the Python code in any of these models using the Advanced Plugin Model Editor. Note that this is actually the same tool as the Python Shell-Editor Tool .
For details of the SasView plugin model format see Writing a Plugin Model .
Note
Model files generated with the Sum/Multi option are still using the SasView 3.x model format. Unless you are confident about what you are doing, it is recommended that you only modify lines denoted with the ## <—– comments!
When editing is complete, select Run > Check Model from the Advanced Plugin Model Editor menu bar. An Info box will appear with the results of the compilation and model unit tests. The model will only be usable if the tests ‘pass’.
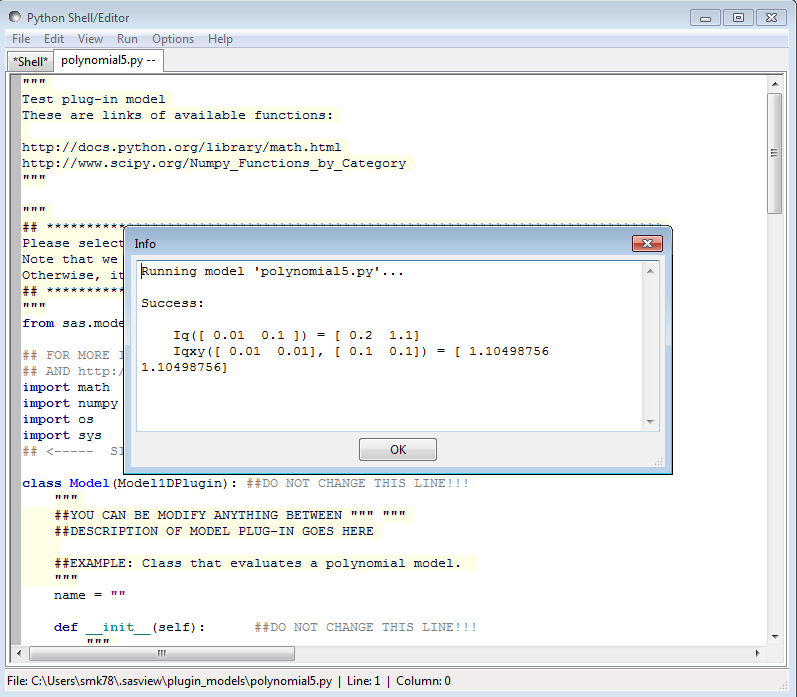
To use the model, go to the relevant Fit Page, select the Plugin Models category and then select the model from the drop-down menu.
Any changes to a plugin model generated in this way only become effective after it is re-selected from the model drop-down menu on the FitPage.
Delete Plugin Models
Simply highlight the plugin model to be removed. The operation is final!!!
NB: Models shipped with SasView cannot be removed in this way.
Load Plugin Models
This option loads (or re-loads) all models present in the ~\.sasview\plugin_models folder.
Fitting Options
It is possible to specify which optimiser SasView should use to fit the data, and to modify some of the configurational parameters for each optimiser.
From Fitting in the menu bar select Fit Options, then select one of the following optimisers:
- DREAM
- Levenberg-Marquardt
- Quasi-Newton BFGS
- Differential Evolution
- Nelder-Mead Simplex
The DREAM optimiser is the most sophisticated, but may not necessarily be the best option for fitting simple models. If uncertain, try the Levenberg-Marquardt optimiser initially.
These optimisers form the Bumps package written by P Kienzle. For more information on each optimiser, see the Fitting Documentation.
Fitting Integer Parameters
Most of the parameters in SasView models will naturally take floating point (decimal) values, but there are some parameters which can only have integer values. Examples include, but are not limited to, the number of shells in a multilayer vesicle, the number of beads in a pearl necklace, the number of arms of a star polymer, and so on. Wherever possible/recognised, the integer nature of a parameter is specified in the respective model documentation and/or parameter table, so read the documentation carefully!
Integer parameters must be fitted with care.
Start with your best possible guess for the value of the parameter. And using a priori knowledge, fix as many of the other parameters as possible.
The SasView optimizers treat integer parameters internally as floating point numbers, but the values presented to the user are truncated or rounded, as appropriate.
In most instances integer parameters will probably be greater than zero. A good policy in such cases is to use a constraint to enforce this.
Because an integer parameter should, by definition, only move in integer steps, problems may be encountered if the optimizer step size is too small. Similarly, be very careful about applying polydispersity to integer parameters.
The Levenberg-Marquardt and Quasi-Newton BFGS (and other derivative-based) optimizers are probably best avoided for fitting models with integer parameters.
Fitting Limits
By default, SasView will attempt to model fit the full range of the data; ie, across all Q values. If necessary, however, it is possible to specify only a sub-region of the data for fitting.
In a FitPage or BatchPage change the Q values in the Min and/or Max text boxes. Vertical coloured bars will appear on the graph with the data and ‘theory’ indicating the current Q limits (red = Qmin, purple = Qmax).
To return to including all data in the fit, click the Reset button.
Shortcuts
Copy/Paste Parameters
It is possible to copy the parameters from one Fit Page and to paste them into another Fit Page using the same model.
To copy parameters, either:
- Select Edit -> Copy Params from the menu bar, or
- Use Ctrl(Cmd on Mac) + Left Mouse Click on the Fit Page.
To paste parameters, either:
- Select Edit -> Paste Params from the menu bar, or
- Use Ctrl(Cmd on Mac) + Shift + Left-click on the Fit Page.
If either operation is successful a message will appear in the info line at the bottom of the SasView window.
Bookmark
To Bookmark a Fit Page either:
- Select a Fit Page and then click on Bookmark in the tool bar, or
- Right-click and select the Bookmark in the popup menu.
Status Bar & Console
The status bar is located at the bottom of the SasView window and displays messages, hints, warnings and errors.
At the right-hand side of the status bar is a button marked Console. The Console displays available message history and some run-time traceback information.
During a long task the Console can also be used to monitor the progress.
Single Fit Mode
NB: Before proceeding, ensure that the Single Mode radio button at the bottom of the Data Explorer is checked (see the section Loading Data ).
This mode fits one data set.
When data is sent to the fitting it is plotted in a graph window as markers.
If a graph does not appear, or a graph window appears but is empty, then the data has not loaded correctly. Check to see if there is a message in the Status Bar & Console or in the Console window.
Assuming the data has loaded correctly, when a model is selected a green model calculation (or what SasView calls a ‘Theory’) line will appear in the earlier graph window, and a second graph window will appear displaying the residuals (the difference between the experimental data and the theory) at the same X-data values. See Assessing Fit Quality.
The objective of model-fitting is to find a physically-plausible model, and set of model parameters, that generate a theory that reproduces the experimental data and minimizes the values of the residuals.
Change the default values of the model parameters by hand until the theory line starts to represent the experimental data. Then check the tick boxes alongside the ‘background’ and ‘scale’ parameters. Click the Fit button. SasView will optimise the values of the ‘background’ and ‘scale’ and also display the corresponding uncertainties on the optimised values.
Note
If the uncertainty on a fitted parameter is unrealistically large, or if it displays as NaN, the model is most likely a poor representation of the data, the parameter in question is highly correlated with one or more of the other fitted parameters, or the model is relatively insensitive to the value of that particular parameter.
In the bottom left corner of the Fit Page is a box displaying a normalised value of the statistical \(\chi^2\) parameter (the reduced \(\chi^2\), See Assessing Fit Quality) returned by the optimiser.
Now check the box for another model parameter and click Fit again. Repeat this process until all relevant parameters are checked and have been optimised. As the fit of the theory to the experimental data improves, the value of ‘Reduced Chi2’ will decrease. A good model fit should produce values of Reduced Chi2 close to one, and certainly << 100. See Assessing Fit Quality.
SasView has a number of different optimisers (see the section Fitting Options). The DREAM optimiser is the most sophisticated, but may not necessarily be the best option for fitting simple models. If uncertain, try the Levenberg-Marquardt optimiser initially.
Simultaneous Fit Mode
NB: Before proceeding, ensure that the Single Mode radio button at the bottom of the Data Explorer is checked (see the section Loading Data ).
This mode is an extension of the Single Fit Mode that allows for some relatively extensive constraints between fitted parameters in a single FitPage or between several FitPage‘s (eg, to constrain all fitted parameters to be the same in a contrast series of FitPages except for the solvent sld parameter, constrain the length to be twice that of the radius in a single FitPage, fix the radius of the sphere in one FitPage to be the same as the radius of the cylinder in a second FitPage, etc).
If the data to be fit are in multiple files, load each file, then select each file in the Data Explorer, and Send To Fitting. If multiple data sets are in one file, load that file, Unselect All Data, select just those data sets to be fitted, and Send To Fitting. Either way, the result should be that for n data sets you have 2n graphs (n of the data and model fit, and n of the resulting residuals). But it may be helpful to minimise the residuals plots for clarity. Also see Assessing Fit Quality.
NB: If you need to use a custom Plugin Model, you must ensure that model is available first (see Adding your own Models ).
Method
Now go to each FitPage in turn and:
Select the required category and model;
Unselect all the model parameters;
Enter some starting guesses for the parameters;
Enter any parameter limits (recommended);
Select which parameters will refine (selecting all is generally a bad idea...);
When done, select Constrained or Simultaneous Fit under Fitting in the menu bar.
In the Const & Simul Fit page that appears, select which data sets are to be simultaneously fitted (this will probably be all of them or you would not have loaded them in the first place!).
To tie parameters between the data sets with constraints, check the ‘yes’ radio button next to Add Constraint? in the Fit Constraints box.
To constrain all identically named parameters to fit simultaneously to the same value across all the Fitpages use the Easy Setup drop-down buttons in the Const & Simul Fit page.
NB: You can only constrain parameters that are set to refine.
Constraints will generally be of the form
Mi Parameter1 = Mj.Parameter1
however the text box after the ‘=’ sign can be used to adjust this relationship; for example
Mi Parameter1 = scalar * Mj.Parameter1
A ‘free-form’ constraint box is also provided.
Many constraints can be entered for a single fit.
When ready, click the Fit button on the Const & Simul Fit page, NOT the Fit button on the individual FitPage‘s.
The results of the model-fitting will be returned to each of the individual FitPage‘s.
Note that the Reduced Chi2 value returned is the SUM of the Reduced Chi2 of each fit. To see the Reduced Chi2 value for a specific FitPage, click the Compute button at the bottom of that FitPage to recalculate. Note that in doing so the degrees of freedom will be set to Npts. See Assessing Fit Quality. Moreover in the case of constraints the degrees of freedom are less than one might think due to those constraints.
Batch Fit Mode
NB: Before proceeding, ensure that the Single Mode radio button at the bottom of the Data Explorer is checked (see the section Loading Data ). The Batch Mode button will be used later on!
This mode sequentially fits two or more data sets to the same model. Unlike in simultaneous fitting, in batch fitting it is not possible to constrain fit parameters between data sets.
If the data to be fit are in multiple files, load each file in the Data Explorer. If multiple data sets are in one file, load just that file. Unselect All Data, then select a single initial data set to be fitted. Fit that selected data set as described above under Single Fit Mode.
NB: If you need to use a custom Plugin Model, you must ensure that model is available first (see Adding your own Models ).
Method
Now Select All Data in the Data Explorer, check the Batch Mode radio button at the bottom of that panel and Send To Fitting. A BatchPage will be created.
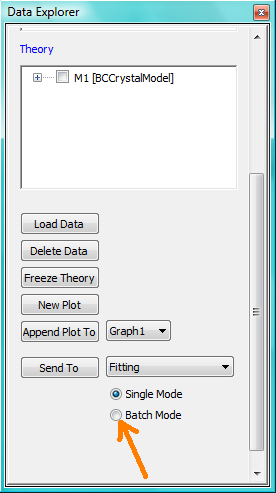
NB: The Batch Page can also be created by checking the Batch Mode radio button and selecting New Fit Page under Fitting in the menu bar.
Using the drop-down menus in the BatchPage, now set up the same data set with the same model that you just fitted in single fit mode. A quick way to set the model parameter values is to just copy them from the earlier Single Fit. To do this, go back to the Single Fit FitPage, select Copy Params under Edit in the menu bar, then go back to the BatchPage and Paste Params.
When ready, use the Fit button on the BatchPage to perform the fitting, NOT the Fit button on the individual FitPage‘s.
Unlike in single fit mode, the results of batch fits are not returned to the BatchPage. Instead, a spreadsheet-like Grid Window will appear.
If you want to visually check a graph of a particular fit, click on the name of a Data set in the Grid Window and then click the View Fits button. The data and the model fit will be displayed. If you select mutliple data sets they will all appear on one graph.
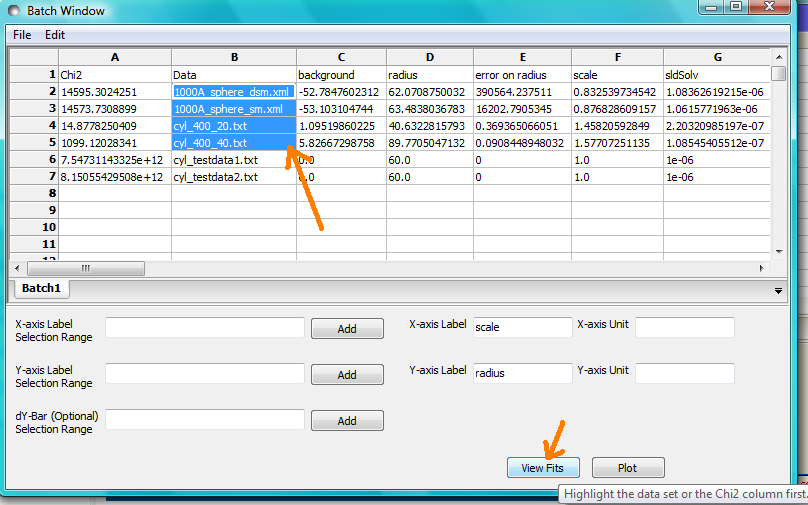
NB: In theory, returning to the BatchPage and changing the name of the I(Q) data source should also work, but at the moment whilst this does change the data set displayed it always superimposes the ‘theory’ corresponding to the starting parameters.
If you select a ‘Chi2’ value and click the View Fits button a graph of the residuals for that data set is displayed. Again, if you select multiple ‘Chi2’ values then all the residuals data will appear on one graph. Also see Assessing Fit Quality.
Chain Fitting
By default, the same parameter values copied from the initial single fit into the BatchPage will be used as the starting parameters for all batch fits. It is, however, possible to get SasView to use the results of a fit to a preceding data set as the starting parameters for the next fit in the sequence. This variation of batch fitting is called Chain Fitting, and will considerably speed up model-fitting if you have lots of very similar data sets where a few parameters are gradually changing. Do not use chain fitting on disparate data sets.
To use chain fitting, select Chain Fitting under Fitting in the menu bar. It toggles on/off, so selecting it again will switch back to normal batch fitting.
Grid Window
The Grid Window provides an easy way to view the results from batch fitting. It will be displayed automatically when a batch fit completes, but may be opened at any time by selecting Show Grid Window under View in the menu bar.
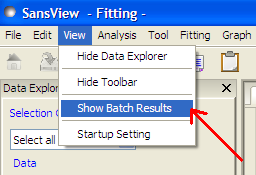
Once a batch fit is completed, all model parameters are displayed but not their uncertainties. To view the uncertainties, click on a given column then go to Edit in the menu bar, select Insert Column Before and choose the required data from the list. An empty column can be inserted in the same way.
To remove a column from the grid, click on the column header and choose Remove Column under Edit in the menu bar. The same functionality also allows you to re-order columns.
NB: You cannot insert/remove/re-order the rows in the Grid Window.
All of the above functions are also available by right-clicking on a column label.
NB: If there is an existing Grid Window and another batch fit is performed, an additional ‘Table’ tab will be added to the Grid Window.
The parameter values in the currently selected table of the Grid Window can be output to a CSV file by choosing Save As under File in the (Grid Window) menu bar. The default filename includes the date and time that the batch fit was performed.
Saved CSV files can be reloaded by choosing Open under File in the Grid Window menu bar. The loaded parameters will appear in a new table tab.
NB: Saving the Grid Window does not save any experimental data, residuals or actual model fits. Consequently if you reload a saved CSV file the ability to View Fits will be lost.
Parameter Plots
Any column of numeric parameter values can be plotted against another using the Grid Window. Simply select one column at the time and click the Add button next to the required X/Y-axis Selection Range text box. When both the X and Y axis boxes have been completed, click the Plot button.
When the Add button is clicked, SasView also automatically completes the X/Y-axis Label text box with the heading from Row 1 of the selected table, but different labels and units can be entered manually.
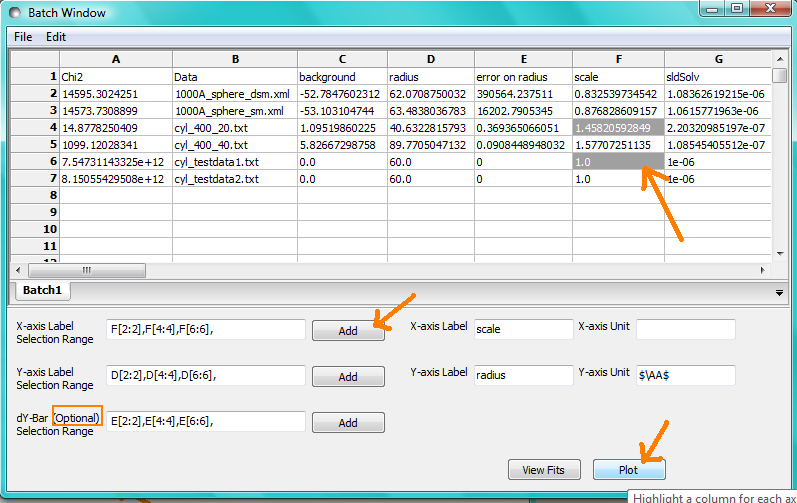
The X/Y-axis Selection Range can be edited manually. The text control box recognises the operators +, -, *, /, or ‘pow’, and allows the following types of expression :
if an axis label range is a function of 1 or more columns, write this type of expression
constant1 * column_name1 [minimum row index : maximum row index] operator constant2 * column_name2 [minimum row index : maximum row index]
Example: radius [2 : 5] -3 * scale [2 : 5]
if only some values of a given column are needed but the range between the first row and the last row used is not continuous, write this type of expression
column_name1 [minimum row index1 : maximum row index1] , column_name1 [minimum row index2 : maximum row index2]
Example: radius [2 : 5] , radius [10 : 25]
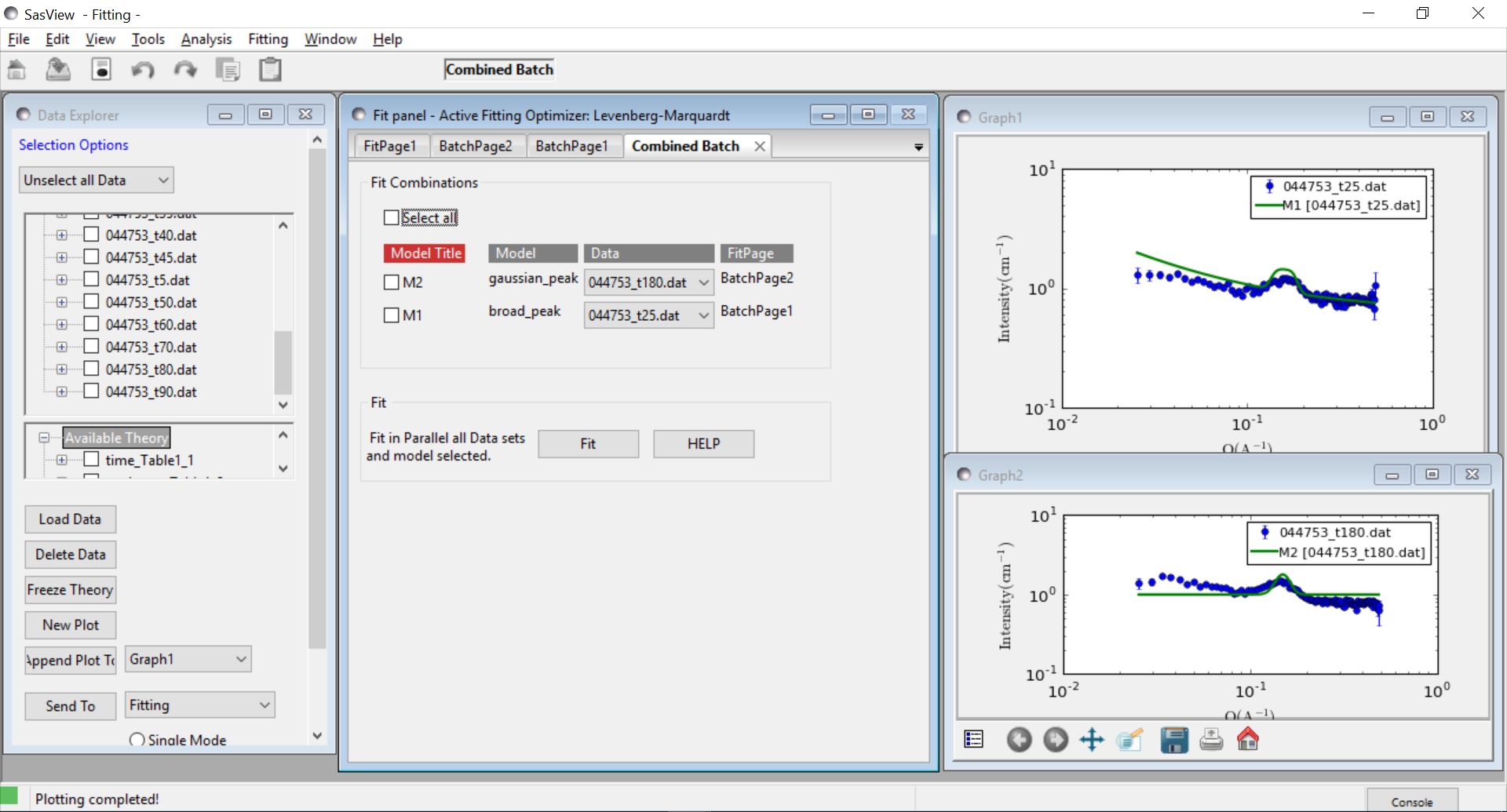
Combined Batch Fit Mode
The purpose of the Combined Batch Fit is to allow running two or more batch fits in sequence without overwriting the output table of results. This may be of interest for example if one is fitting a series of data sets where there is a shape change occurring in the series that requires changing the model part way through the series; for example a sphere to rod transition. Indeed the regular batch mode does not allow for multiple models and requires all the files in the series to be fit with single model and set of parameters. While it is of course possible to just run part of the series as a batch fit using model one followed by running another batch fit on the rest of the series with model two (and/or model three etc), doing so will overwrite the table of outputs from the previous batch fit(s). This may not be desirable if one is interested in comparing the parameters: for example the sphere radius of set one and the cylinder radius of set two.
Method
In order to use the Combined Batch Fit, first load all the data needed as described in Loading Data. Next start up two or more BatchPage fits following the instructions in Batch Fit Mode but DO NOT PRESS FIT. At this point the Combine Batch Fit menu item under the Fitting menu should be active (if there is one or no BatchPage the menu item will be greyed out and inactive). Clicking on Combine Batch Fit will bring up a new panel, similar to the Const & Simult Fit panel. In this case there will be a checkbox for each BatchPage instead of each FitPage that should be included in the fit. Once all are selected, click the Fit button on the BatchPage to run each batch fit in sequence
The batch table will then pop up at the end as for the case of the simple Batch Fitting with the following caveats:
Note
The order matters. The parameters in the table will be taken from the model used in the first BatchPage of the list. Any parameters from the second and later BatchPage s that have the same name as a parameter in the first will show up allowing for plotting of that parameter across the models. The other parameters will not be available in the grid.
Note
a corralary of the above is that currently models created as a sum|multiply model will not work as desired because the generated model parameters have a p#_ appended to the beginning and thus radius and p1_radius will not be recognized as the same parameter.
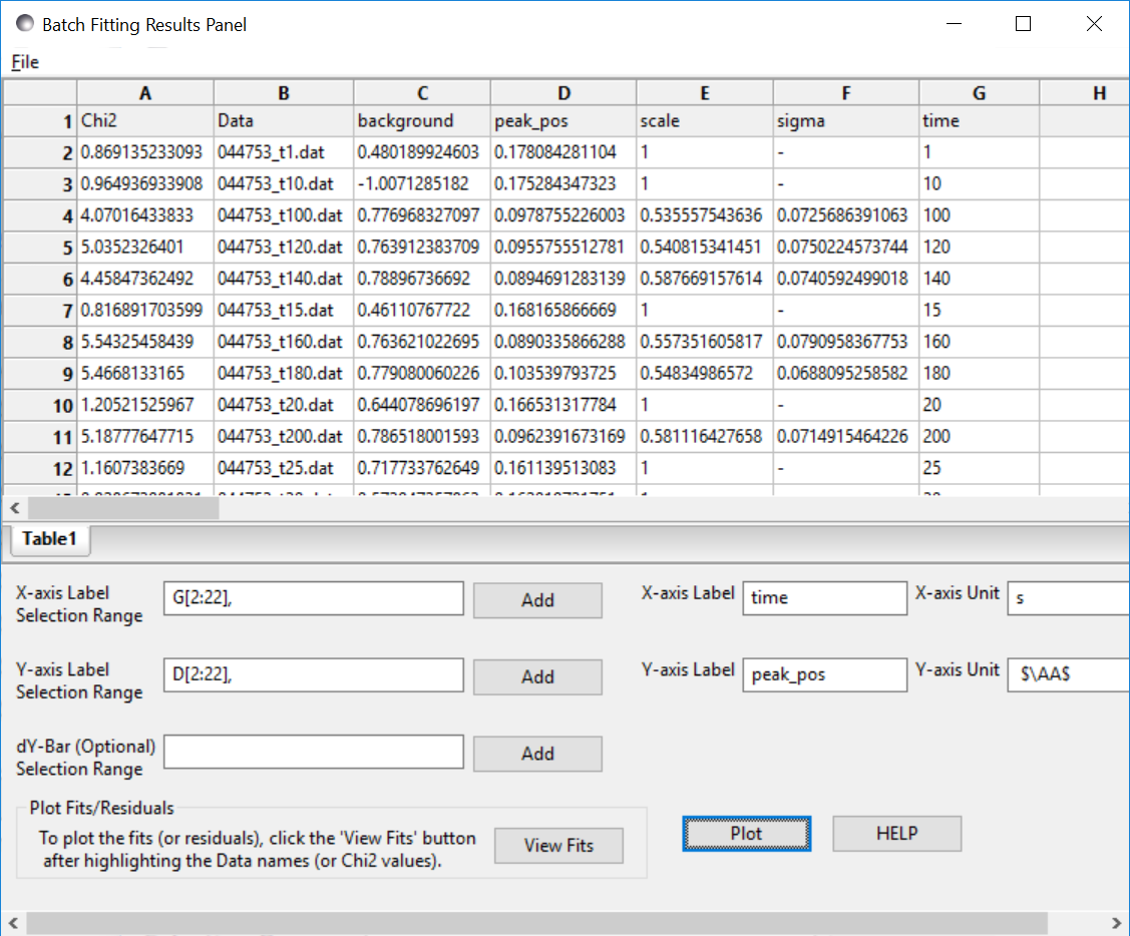
In the example shown above the data is a time series with a shifting peak. The first part of the series was fitted using the broad_peak model, while the rest of the data were fit using the gaussian_peak model. Unfortunately the time is not listed in the file but the file name contains the information. As described in Grid Window, a column can be added manually, in this case called time, and the peak position plotted against time.
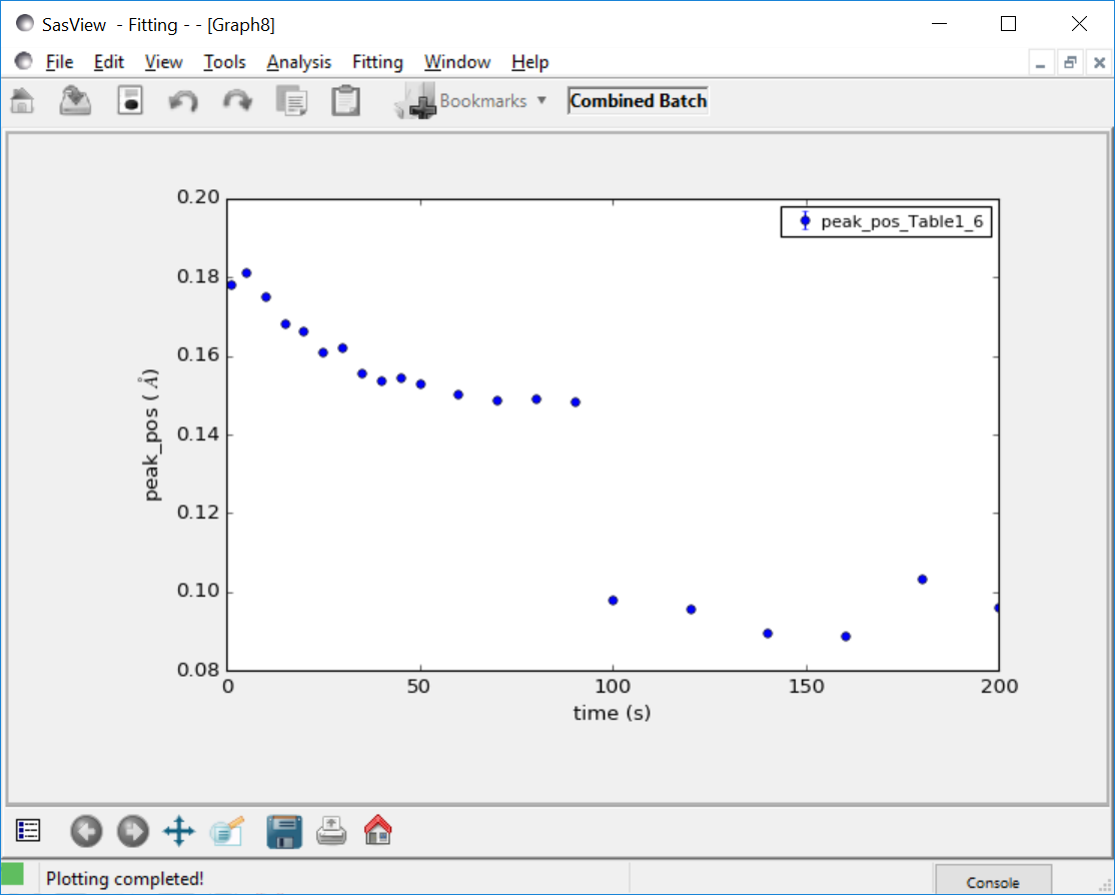
Note the discontinuity in the peak position. This reflects the fact that the Gaussian fit is a rather poor model for the data and is not actually finding the peak.
.*Document History*